Cytoscape is a popular open source platform for visualising molecular interaction networks.
Cytoscape websiteCytoscape has a built-in plugin manager (Plugins -> Manage Plugins) which lets the user install a large number of plugins.
One of the most important applications of Cytoscape is the analysis of interaction networks. The networks are described by eXtensible Graph Markup and Modeling Language - XGMML.
XGMMLI found a sample file here
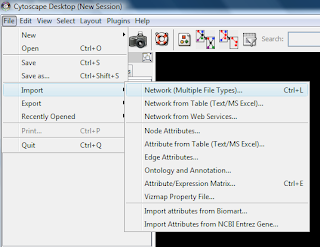
pte.xgmmlTo import the file into Cytoscape, select File -> Import -> Network(multiple file types)
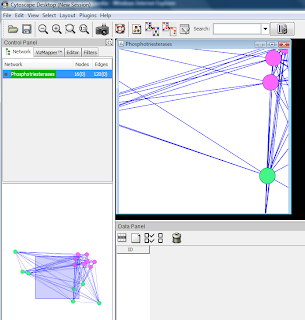
This is how a network typically may look.
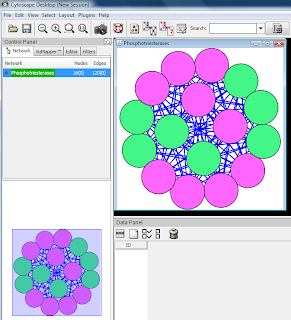
To view all nodes and edges, apply a layout, for example Layouts -> Cytoscape Layouts -> Spring Embedded
In the Node Attribute Browser below, click "Select All Attributes". To select all nodes in the Cytoscape model, use Ctrl-A.
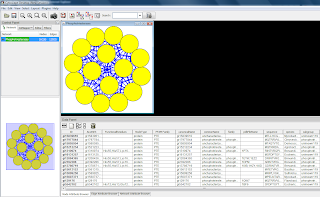
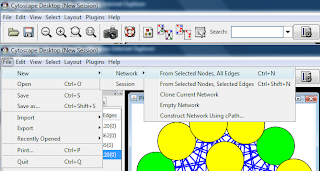
If the network is large, it is possible to use a selection criteria to select nodes of interes and to create a new network from the selected nodes only. For example, it is possible to sort the nodes by an attribute in the node attribute browser (or edge attribute browser), select a number of nodes and then select File -> New -> Network -> From Selected Nodes, All Edges. Cytoscape will create a child subnetwork which will only include selected nodes and/or edges.
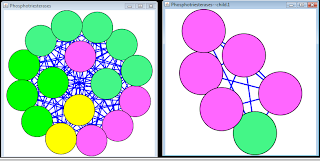
by Evgeny. Also posted on my website
No comments:
Post a Comment